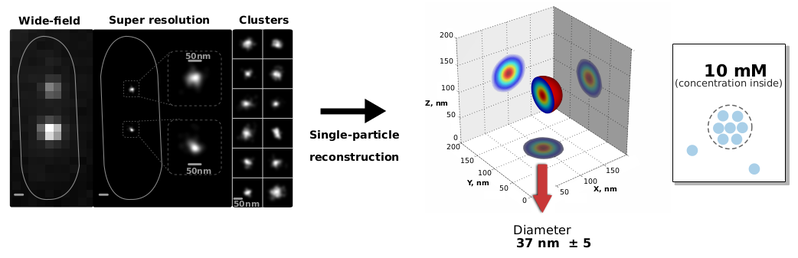
Who we areWelcome to Marcelo Nollmann's research group at the Center for Structural Biochemistry of the CNRS/INSERM We develop single-molecule and advanced microscopy methodologies to understand the mechanisms underlying DNA organization and transcription in multi-cellular organisms. Click here for links to our multiplexed FISH (Hi-M) acquisition (qudi-CBS) and analysis (pyHiM) software packages.News
|
|
Featured Publications
Multiple parameters shape the 3D chromatin structure of single nuclei
|
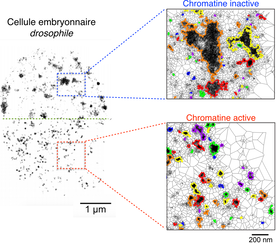
3D chromatin interactions involving Drosophila insulators are infrequent but preferential and arise before TADs and transcription
|
|
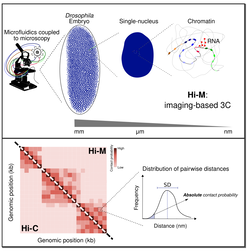
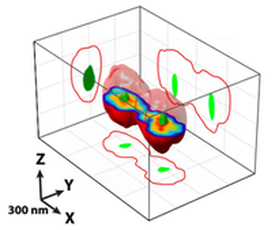
Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms
Cardozo-Gizzi, et al. Mol Cell (2019) |
Direct and simultaneous observation of transcription and chromosome architecture in single cells with Hi-M.
Cardozo-Gizzi, et al. Nature Protocols (2020) |
A complete list of publications, including chapters and articles not accessed by PubMed, can be found here.