Single-molecule and super-resolution
An important part of our group works on the development of single-molecule and super resolution methods. In particular, we develop spatial genomics, single-molecule localization microscopy using adaptive optics, implement variants of structured illumination microscopy, and work on the development of multi-focus microscopes. These implementations are combined with other methods such as microfluidics or DNA origami.
Spatial genomics and transcriptomics
Our current technology development is focused on imaging-based spatial genomics methods. All publications are available in our Github repository.
See these recent publications for more information:
Direct and simultaneous observation of transcription and chromosome architecture in single cells with Hi-M.
Andrés M. Cardozo Gizzi, Sergio M. Espinola, Julian Gurgo, Christophe Houbron, Jean-Bernard Fiche, Diego I. Cattoni, Marcelo Nollmann
Nature Protocols 15(3):840-876. doi: 10.1038/s41596-019-0269-9. Epub 2020 Jan 22.
See full updated protocol in our Hi-M Github page.
Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms.
AM Cardozo Gizzi, D I. Cattoni, J-B Fiche, S Espinola, J Gurgo, O Messina, C Houbron, Y Ogiyama, G-L Papadopoulos, G Cavalli, M Lagha, M Nollmann
Molecular Cell, 2019 Apr 4;74(1):212-222.e5. doi: 10.1016/j.molcel.2019.01.011. Epub 2019 Feb 19. [BioRxiv pre-print]
Comment on our article by Daniel Larson here.
Recommended by F1000: Martí-Renom M: F1000Prime Recommendation of [Cardozo Gizzi AM et al., Mol Cell 2019 74(1):212-222.e5]. In F1000Prime, 19 Aug 2019; 10.3410/f.735120435.793559990
Single-molecule localization microscopy
In the past, we developed several flavors of single-molecule localization microscopy (SMLM). These include
Principles of super-resolution microscopy
Components of our basic SMLM microscope
Super-resolution imaging of bacteria in a micro-fluidics device.
Cattoni* DI, Fiche* JB, Valeri A, Mignot T, Nöllmann M.
PLoS ONE 8(10):e76268, Oct 2013. [PDF]
Single-molecule super-resolution imaging in bacteria
Diego I. Cattoni, Jean-Bernard Fiche, Marcelo Nöllmann
Current Opinion in Microbiology, 15(6):758-63, Dec 2012. [PDF]
Nanometer resolved single-molecule colocalization of nuclear factors by two-color super resolution microscopy imaging
Mariya Georgieva; Diego I Cattoni; Jean-Bernard Fiche; Thibaut Mutin; Delphine Chamousset; Marcelo Nollmann. Methods pii: S1046-2023(16)30063-9. 1 Apr 2016 . [PDF]
Our current technology development is focused on imaging-based spatial genomics methods. All publications are available in our Github repository.
See these recent publications for more information:
Direct and simultaneous observation of transcription and chromosome architecture in single cells with Hi-M.
Andrés M. Cardozo Gizzi, Sergio M. Espinola, Julian Gurgo, Christophe Houbron, Jean-Bernard Fiche, Diego I. Cattoni, Marcelo Nollmann
Nature Protocols 15(3):840-876. doi: 10.1038/s41596-019-0269-9. Epub 2020 Jan 22.
See full updated protocol in our Hi-M Github page.
Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms.
AM Cardozo Gizzi, D I. Cattoni, J-B Fiche, S Espinola, J Gurgo, O Messina, C Houbron, Y Ogiyama, G-L Papadopoulos, G Cavalli, M Lagha, M Nollmann
Molecular Cell, 2019 Apr 4;74(1):212-222.e5. doi: 10.1016/j.molcel.2019.01.011. Epub 2019 Feb 19. [BioRxiv pre-print]
Comment on our article by Daniel Larson here.
Recommended by F1000: Martí-Renom M: F1000Prime Recommendation of [Cardozo Gizzi AM et al., Mol Cell 2019 74(1):212-222.e5]. In F1000Prime, 19 Aug 2019; 10.3410/f.735120435.793559990
Single-molecule localization microscopy
In the past, we developed several flavors of single-molecule localization microscopy (SMLM). These include
- Improved setup for two-color STORM with 5nm registration precision
- 3D-SMLM microscope with adaptive optics for aberration correction and user-defined astigmatism
- 2D-SMLM coupled to atomic force microscopy
- Co-localization analysis from two-color STORM data
Principles of super-resolution microscopy
Components of our basic SMLM microscope
Super-resolution imaging of bacteria in a micro-fluidics device.
Cattoni* DI, Fiche* JB, Valeri A, Mignot T, Nöllmann M.
PLoS ONE 8(10):e76268, Oct 2013. [PDF]
Single-molecule super-resolution imaging in bacteria
Diego I. Cattoni, Jean-Bernard Fiche, Marcelo Nöllmann
Current Opinion in Microbiology, 15(6):758-63, Dec 2012. [PDF]
Nanometer resolved single-molecule colocalization of nuclear factors by two-color super resolution microscopy imaging
Mariya Georgieva; Diego I Cattoni; Jean-Bernard Fiche; Thibaut Mutin; Delphine Chamousset; Marcelo Nollmann. Methods pii: S1046-2023(16)30063-9. 1 Apr 2016 . [PDF]
Multi-focus microscopy
|
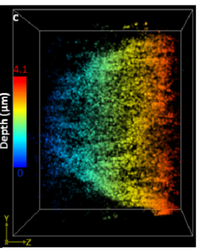
We develop new microscopies based on multi-focus microscopy (Abrahamsson, 2013). In particular, we focus on the development and construction of MFG gratings in collaboration with the LAAS (CNRS, Toulouse). Recently, we developed a 3D super-resolution microscopy method that enables deep imaging in cells based on a combination of Multifocus Microscopy and PSF engineering. We demonstrated the performance of the method by imaging the nuclear envelope of eukaryotic cells reaching a depth of field of ~ 4μm. Also, we improved the efficiency of transmission of MFGs by using multi-phase engraving.
Astigmatic multifocus microscopy enables deep 3D super-resolved imaging. Laura Oudjedi, Jean-Bernard Fiche, Sara Abrahamsson, Laurent Mazenq, Aurélie Lecestre, Pierre-François Calmon, Aline Cerf, and Marcelo Nöllmann Biomedical Optics Express, 2016, in press. [PDF] Perfect color Multifocus Microscopy (MFM) with increased sensitivity Sara Abrahamsson, Rob Ilic, Jan Wisniewski, Brian Mehl, Liya Yu, Lei Chen, Marcelo Davanco, Laura Oudjedi, Jean-Bernard Fiche, Bassam Hajj, Xin Jin, Joan Pulupa, Christine Cho, Mustafa Mir, Xavier Darzacq, Marcelo Nollmann, Maxime Dahan, Carl Wu, Timothy Lionett, James . Liddle and Cornelia I. Bargmann Biomedical Optics Express, 2016, in press |
Single-molecule manipulation/fluorescence
|
Magnetic Tweezers coupled to single-molecule fluorescence detection
We developed a standard magnetic tweezers coupled to TIRF illumination and single-molecule fluorescence detection to monitor the activity, mechanical properties, and position of fluorescently-labelled proteins in real-time. The setup has been used for the study of SpoIIIE (Thakur, submitted) and the mechanism of looping of insulator proteins. The fluorescence properties and binding mechanism of SYTOX green, a bright, low photo-damage DNA intercalating agent. Thakur, S,Cattoni DI, Nollmann M European Biophysical Journal, July 2015, 44(5):337-48. [PDF] Direct observation of the translocation mechanism of transcription termination factor Rho Gocheva V, Le Gall A, Boudvillain M, Margeat E, Nollmann M Nucleic Acids Research, 2015 Feb 27; 43(4):2367-77. [PDF] Constructing a magnetic tweezers to monitor RNA translocation at the single-molecule level. Salas D, Gocheva V, Nöllmann M. Methods Mol Biol. 1259:257-73. Jan 2015. [PDF] |