Principles of SMLM
The image that a point source makes on a camera is called the point-spread function (PSF). The PSF is limited by diffraction to be no less than approximately half the wavelength of light. Until very recently, the spread of the PSF defined the maximum resolution attainable by conventional fluorescence microscopy. Several methods have been recently developed that circumvent this limit by oversampling the PSF and locating its center with nanometer accuracy: single-molecule high-resolution localization (SHREC; Churchman and Spudich, 2005), fluorescence imaging with one nanometer accuracy (FIONA; Yildiz and Selvin, 2005), photo-activated localization (PALM; Betzig, 2006 and FPALM; Hess, 2006) and stochastic optical reconstruction microscopy (STORM; Bates, 2006). The basic principles of PALM and STORM are the same and the difference in acronyms is due to historical reasons. For simplicity, we prefer calling the method 'Single-Molecule Super-Resolution Microscopy' or smSRM. In smSRM each fluorophore located in the diffraction-limited spot is photoactivated and localized with nano-meter precision one at a time. The positions of individual positions of fluorescent-labeled proteins or organic dyes composing a molecular complex can be found with nanometer resolution (~20 nm). smSRM usee either genetically encoded photoactivable markers and is thus amenable to live microscopy (Shrof, 2008) or organic dyes.
Because of the need to collect as many photons as possible above background (~500) from each fluorophore in order to localize it with nanometer accuracy, smSRM has been generally used with a total internal reflection fluorescence microscope (penetration depth ~100 nm) due to its high background rejection. However, in order to localize molecular complexes that are deep in the cell (~500 nm) other observation modes still possessing large signal to noise ratios need to be implemented. See Figure on the bottom for a further schematic description of the conceptual principles of smSRM.
Because of the need to collect as many photons as possible above background (~500) from each fluorophore in order to localize it with nanometer accuracy, smSRM has been generally used with a total internal reflection fluorescence microscope (penetration depth ~100 nm) due to its high background rejection. However, in order to localize molecular complexes that are deep in the cell (~500 nm) other observation modes still possessing large signal to noise ratios need to be implemented. See Figure on the bottom for a further schematic description of the conceptual principles of smSRM.
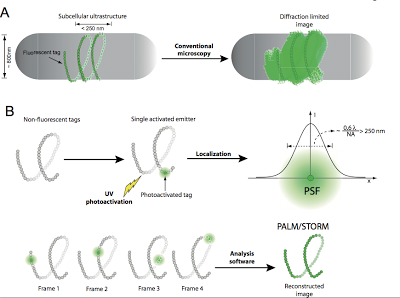
Figure 1. Principles of single-molecule super-resolution microscopy. A- In a conventional microscopy image of a subcellular structure with one of its dimensions smaller than the diffraction limit (~250 nm), all individual fluorophores emit simultaneously and internal structural or dynamical information cannot be resolved. B- smSRM uses photo-switchable or photo-activatable fluorophores. Initially, these fluorophores are in a non-fluorescent state and irradiation with light of the appropriate wavelength induces the stochastic photo-activation of a single fluorophore per diffraction-limited spot. Image analysis is used to automatically detect the fluorophore and its mean position is determined by PSF fitting (upper panel). Sequential cycles of photo-activation/localization/photobleaching are used to obtain the localization of each fluorophore in the structure. These coordinates are finally combined to reconstruct the structure at a resolution that depends on the labeling density and the localization precision with which single emitters were localized. (lower panel).